-
Notifications
You must be signed in to change notification settings - Fork 1
Description
Brief description
Models in computational neuroscience (particularly detailed models with ion channels & multicompartmental cells) are generally based on previous work, with minor or major alterations to the code. Recording the history/relationship of these can be useful for finding out which contributor added which parameters/features and why.
This task aims to make a standard way to record these relationships in structured files and generate graphical representations to elucidate the relationships.
Information
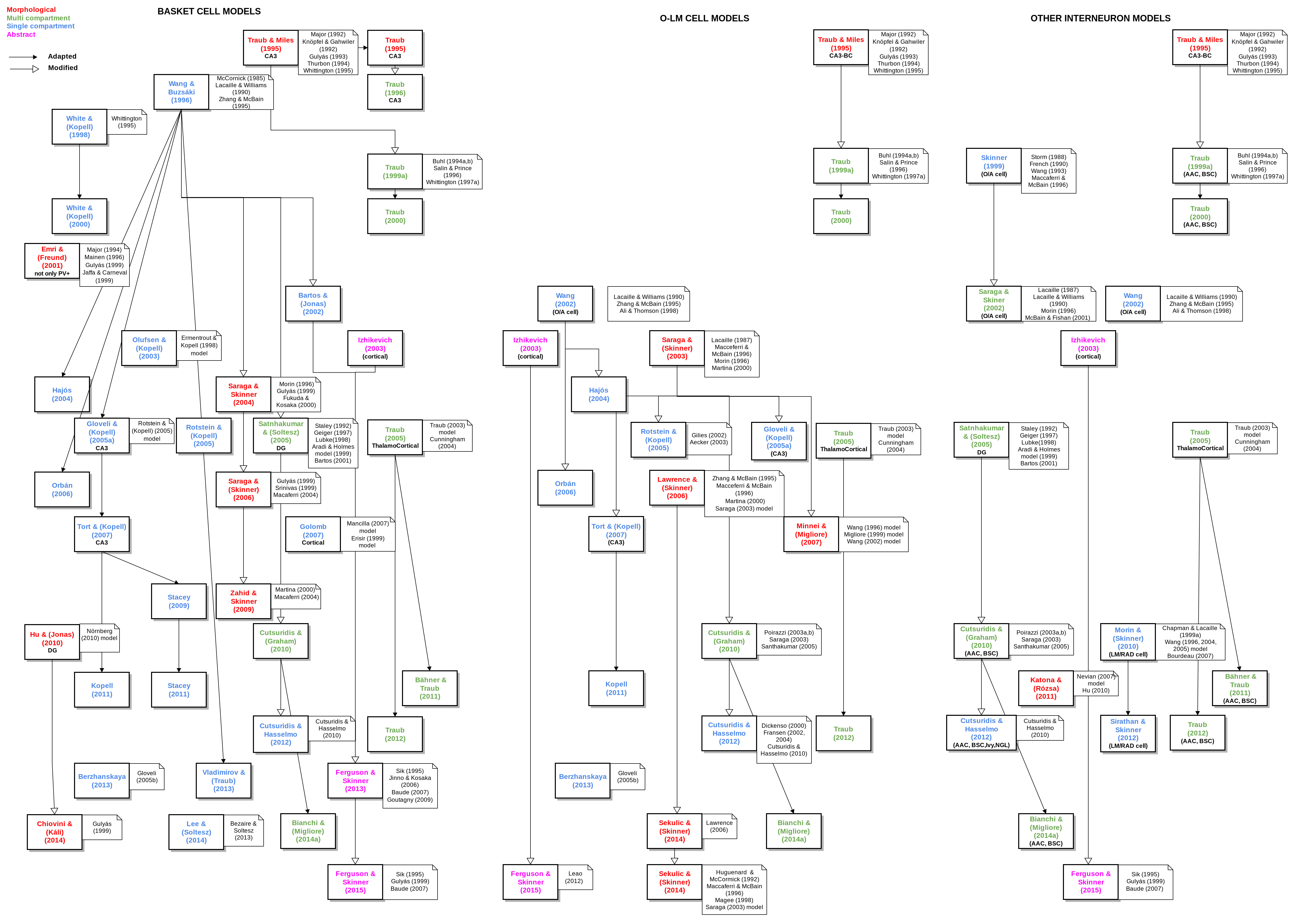
Work on this has started here: https://github.com/OpenSourceBrain/CommunityModellingCA1. One of the aims of this project is to record the various cell/network models of hippocampal region CA1. Manual images of the relationships between modles in this area have been generated by @andrisecker, e.g.
However, it would be better if such diagrams could be generated automatically from structured text descriptions, see here. This would encourage updates to the ancestry description; a researcher would only have to add a line to the text file and the image can be updated.
A script has been started to generate an image like the above from this, see OpenSourceBrain/CommunityModellingCA1#4. This needs to be improved and other output formats supported:
- update python script for generating (nicely formatted) png
- generate interactive/clickable/expandable web page from this using javascript (maybe https://d3js.org/)
Once the format for these csv files is stable and the images/views can be generated other model ancestry files can be requested from the community (cerebellum, cortex, basal gangli, olfactory cortex, etc.)
Knowledge prerequisites
Python
JSON
JavaScript (optional)
Computational neuroscience knowledge useful, but not required.
Contact
Contact p.gleeson -at- ucl.ac.uk if you are interested in contributing to this task, or fork https://github.com/OpenSourceBrain/CommunityModellingCA1 and start working.